
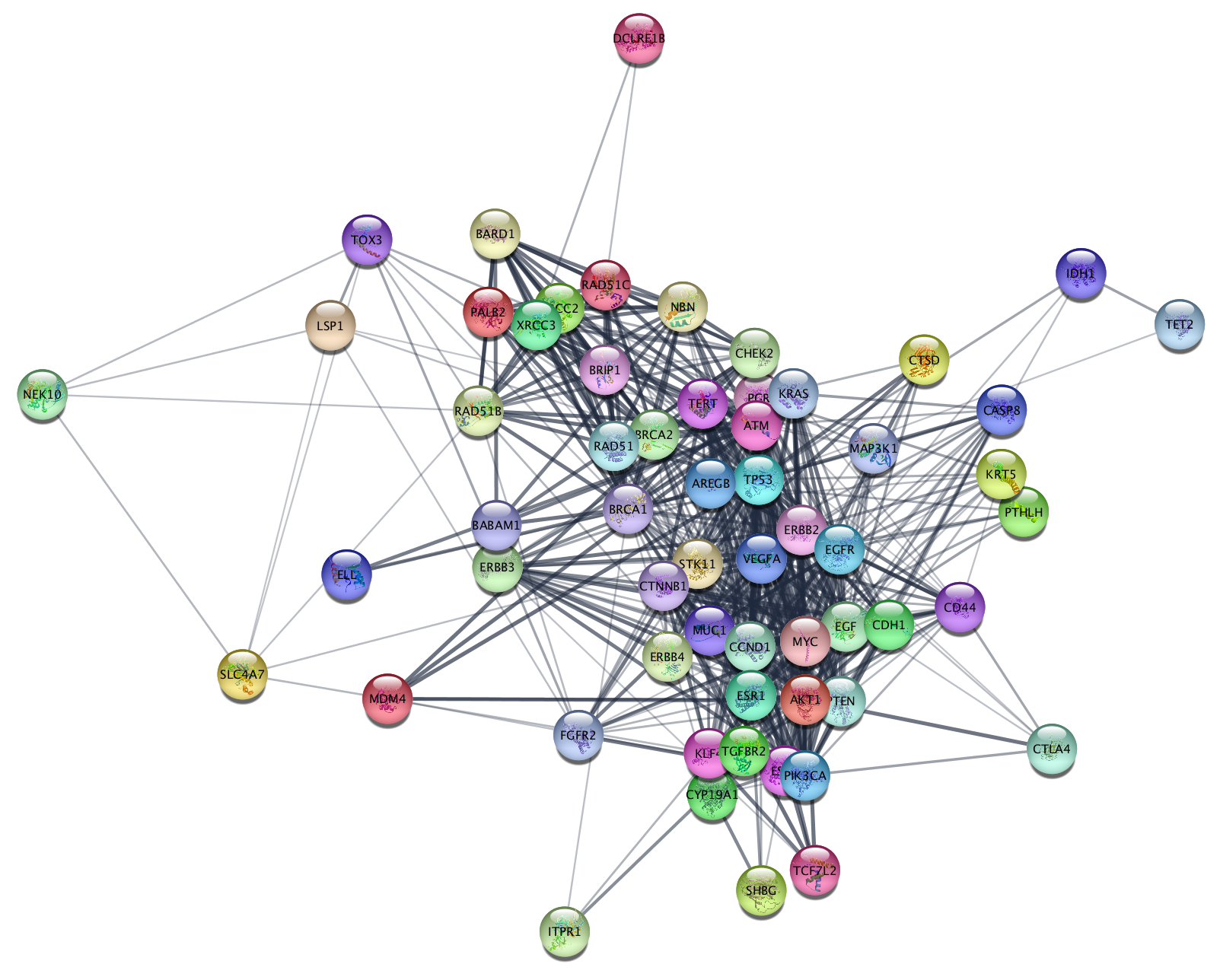
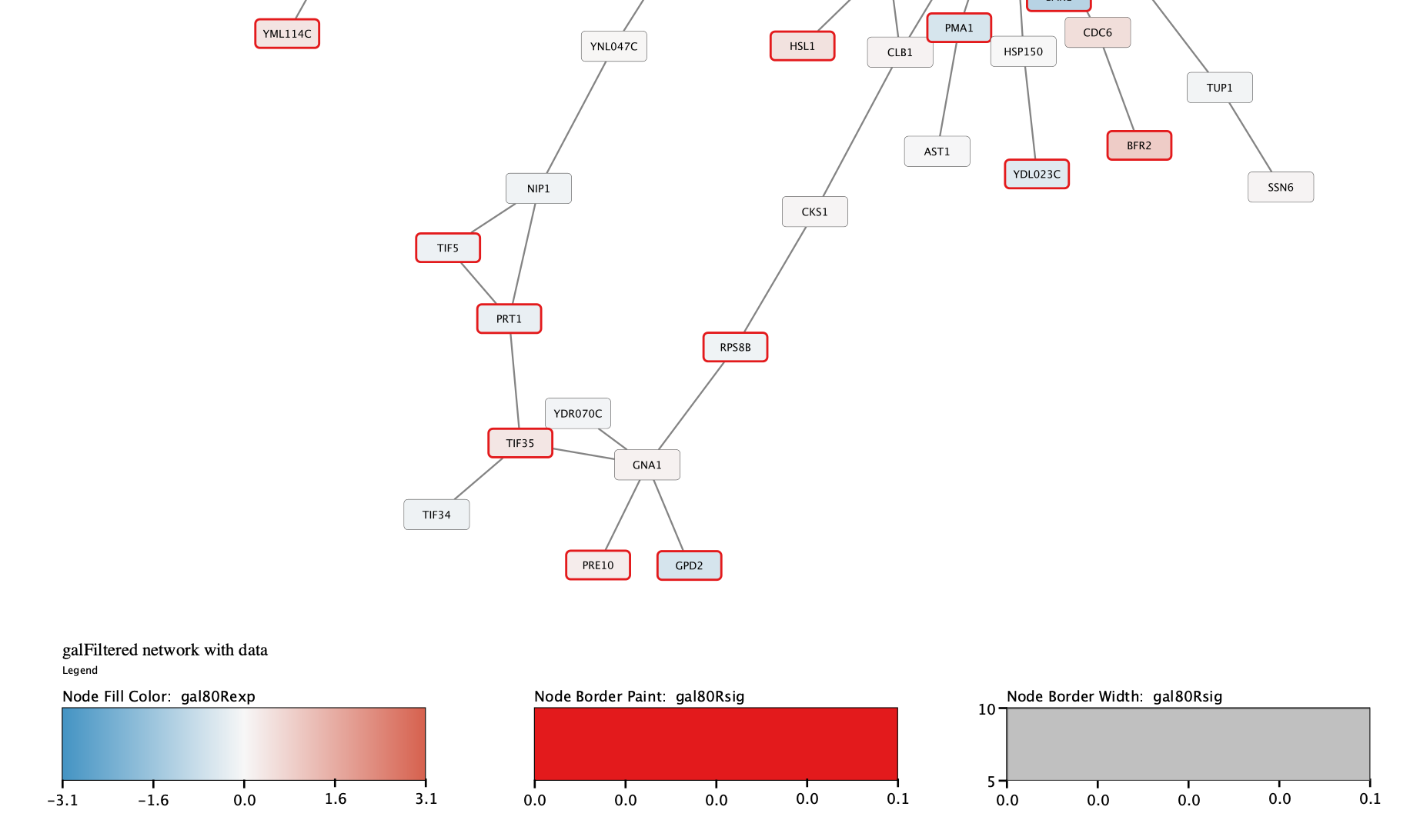
One of the most familiar approaches is the hierarchical clustering of genes and their expression levels under various conditions to produce a dendrogram and heat map (Figure 1A) for analyzing and visualizing microarray data . Clustering algorithms detect patterns within data sets, and organize related genes, proteins, or other key elements to highlight those patterns. One particular technique that has seen wide use in 'omics studies is clustering. Along with this increase in the different types and amount of data, there have been many advances in analytical techniques. High-throughput techniques to generate genomic, proteomic, transcriptomic, metabolomic, and interactomic data continue to advance, generating huge data sets covering more species and more information about the biology of individual species than ever before. clusterMaker is available via the Cytoscape plugin manager. Several of these visualizations and algorithms are only available to Cytoscape users through the clusterMaker plugin. The Cytoscape plugin clusterMaker provides a number of clustering algorithms and visualizations that can be used independently or in combination for analysis and visualization of biological data sets, and for confirming or generating hypotheses about biological function. Cytoscape session files for all three scenarios are provided in the Additional Files section. For scenario three, we explore the possible annotation of a protein as a methylmalonyl-CoA epimerase within the VOC superfamily. For scenario two, we explore the prefoldin complex in detail using both physical and genetic interaction clusters. For scenario one, we explore functionally enriched mouse interactomes specific to particular cellular phenotypes and apply fuzzy clustering. Results are presented in the form of three scenarios of use: analysis of protein expression data using a recently published mouse interactome and a mouse microarray data set of nearly one hundred diverse cell/tissue types the identification of protein complexes in the yeast Saccharomyces cerevisiae and the cluster analysis of the vicinal oxygen chelate (VOC) enzyme superfamily.
CYTOSCAPE SELETION PLUS
clusterMaker is the first Cytoscape plugin to implement such a wide variety of clustering algorithms and visualizations, including the only implementations of hierarchical clustering, dendrogram plus heat map visualization (tree view), k-means, k-medoid, SCPS, AutoSOME, and native (Java) MCL. The Cytoscape network is linked to all of the other views, so that a selection in one is immediately reflected in the others. Here we present clusterMaker, a Cytoscape plugin that implements several clustering algorithms and provides network, dendrogram, and heat map views of the results. For example, protein-protein interaction data sets have been clustered to identify stable complexes, but scientists lack easily accessible tools to facilitate combined analyses of multiple data sets from different types of experiments.
In the post-genomic era, the rapid increase in high-throughput data calls for computational tools capable of integrating data of diverse types and facilitating recognition of biologically meaningful patterns within them.


 0 kommentar(er)
0 kommentar(er)
